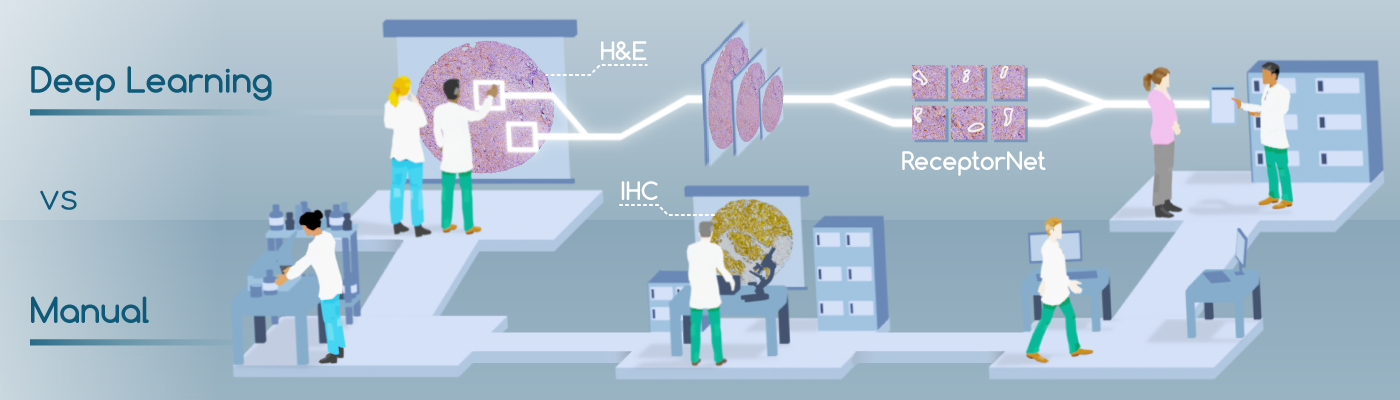
If a woman gets breast cancer, determining the best course of therapy can be quite challenging. The manual process involves taking a biopsy of the tissue, then analyzing its physical structure by staining the tissue with hematoxylin and eosin (H&E)—compounds which attach to tissue morphologies. Next, a pathologist will analyze the tissue’s chemical composition, using immunohistochemistry (IHC) stains, to determine the presence or absence of estrogen and other hormonal receptors. The presence or absence of these receptors will determine the course of therapy. The challenge is that IHC is an expensive, inconsistent, and time-consuming method that is not widely available in developing countries. H&E stains, however, are inexpensive and widely available.
We present ReceptorNet [1], an AI algorithm that determines estrogen receptor status (ERS) from H&E-stained whole slide images of breast cancer tissue, with the accuracy of pathologists analyzing IHC stains. ReceptorNet achieves this by identifying predictive visual features too subtle to be perceived by human eyes. ReceptorNet is trained with clinical data readily available from patient records (with no need for additional manual labels), identifies explainable visual features responsible for decision making, and shows equal performance on data from different countries and demographic groups.
ReceptorNet could improve cancer patient outcomes and clinical accessibility, especially in developing countries. Deployed at scale, ReceptorNet has the potential to deliver world-class oncology decisions in under-resourced settings.
Breast Cancer and Hormone Receptors: Key Concepts
Breast cancer is a major cause of death in women. Two million women were diagnosed with breast cancer in 2018 globally, resulting in 600,000 deaths. A large majority of invasive breast cancers are hormone receptor-positive—the tumor cells grow in the presence of estrogen (ER) and/or progesterone (PR). Patients with hormone-receptor positive tumors are usually prescribed hormone therapies. The US National Comprehensive Cancer Network guidelines mandate that hormone receptor status, including ER status (ERS), be determined for every new patient.
As described above, in clinical workflows, a patient’s tissue sample is stained using hematoxylin and eosin (H&E) staining, followed by visual diagnosis by a pathologist. For positive breast cancer, molecular immunohistochemistry (IHC) is performed as the next step, and ERS is determined by a pathologist by visual inspection of the IHC slide. This process has several limitations: IHC staining is expensive, time-consuming, and not widely available in developing countries. There can be significant variation in IHC sample quality. Finally, the pathologists’ decision-making process is inherently subjective and can result in human errors. These factors lead to variability in ERS determination. An estimated 20% of current IHC-based determinations of ER and PR testing may be inaccurate [2], placing these patients at risk for suboptimal treatment.
ReceptorNet: Determining Hormone Receptor Status using AI
In this paper, we show that the visual appearance of the tumor, captured in the H&E stain, can be directly used to determine estrogen receptor status using AI, without the need to use an IHC stain. We note that the H&E stain does not contain human-identifiable visual features for reliable estimation of ERS, and thus physicians are not trained to perform this task.
Machine learning-driven histopathology methods have relied on expensive, time-consuming, pixel-level pathologist annotations of whole slide images for training. Since pathologists are not clinically trained to determine ERS from H&E stains, they cannot provide manual annotations for these images. Instead, our algorithm is trained with slide-level clinical ERS information already available from patient records and requires no additional pixel-level annotations.
If a tumor has been determined to be ER-negative (ER-) from an IHC stain, we can assume that their H&E WSI contains almost no discriminative features for ER-positivity. Conversely, if the patient has been determined to be ER-positive (ER+), we can assume that at least some regions of their H&E WSI contain discriminative features for ER-positivity. Thus, we can train models using H&E stains as input data, and IHC annotations as input data labels. This problem setup works well with Multiple Instance Learning (MIL)[3]. The goal of MIL is to learn from a training set consisting of labeled “bags” of unlabeled instances. A positively labeled bag contains at least one positive instance, while a negatively labeled bag contains only negative instances. A trained MIL algorithm is able to predict the positive/negative label for unseen bags. We utilize MIL to estimate ERS from a bag of tiles randomly selected from a WSI.

In addition to being accurate, an AI algorithm for ERS estimation should be interpretable—it should enable users to identify regions of the image that are important for decision-making. Interpretability is important for gaining physicians’ trust, for building a robust decision-making system, and to help overcome regulatory concerns. To achieve interpretability, we design an “attention-based” deep neural network that performs MIL [4], which we call ReceptorNet. ReceptorNet learns to assign high attention weights to tiles in the H&E image that are the most important for decision-making, and to assign low attention weights to tiles that are insignificant for this task. We improve on the existing attention-based MIL algorithm using AI techniques including cutout regularization, hard-negative mining and a novel mean pixel regularization.
Training and Evaluating ReceptorNet

To train and test ReceptorNet, we combine two datasets containing a total of 3,399 H&E patient images: the Australian Breast Cancer Tissue Bank (ABCTB) and The Cancer Genome Atlas (TCGA), containing data from three countries. The combined dataset has large variation in preparation and scanning quality. We divide the combined dataset into a train set (2,728 patients) and a test set (671 patients). After performing 5-fold cross-validation on the train set, we train ReceptorNet using all slides from the train set and evaluate it on the test set slides.
We report results using the area under the curve (AUC) for ER+/ER- binary classification. ReceptorNet obtains an AUC of 0.899 on the cross-validation of the train set and an AUC of 0.92 on the test set. On the test set, our method obtains a PPV of .932 and an NPV of .741 at a threshold of .25. Since we do not have access to the original tissue material, it is not possible to perform an exact comparison between pathologists and our approach. But we note that the results from the International Breast Cancer Study Group [2] dataset from a separate study, translate to a PPV of .92 and an NPV of .683 for concordance between primary institution and central testing at the same threshold, comparable to the performance of ReceptorNet.

There is no statistically significant difference (p>0.05, F-test) in ReceptorNet performance across data from Australia, Germany, Poland, and USA. We also do not find statistically significant differences in prediction performance based on patient’s race or menopausal status. In sum, ReceptorNet does not exhibit a bias in performance based on geographic origin or important demographic characteristics of the patient.
Explaining ReceptorNet Decisions

Analyzing attention weights assigned to different regions of an H&E image allows us to determine which tiles were utilized to make ERS estimation. An expert breast cancer pathologist manually reviewed groups of high-attention tiles clustered based on their image features and sorted by their ER+ percentage. ReceptorNet discovers that visual patterns within H&E slides that identify low grade tumors, Invasive Lobular Carcinoma, reactive stroma and in situ carcinoma are predictive of ER-positivity. Prior research has found statistically associations [5-9] between these cancer characteristics and ER+ breast cancers. ReceptorNet also discovers that visual patterns within H&E slides that identify high grade tumors and necrotic debris are predictive of ER-negativity.

Impact and Future Directions
ReceptorNet’s ability to determine IHC-derived hormone receptor status from H&E stains has the potential to improve clinical outcomes and decrease costs in developed countries. In developing countries, where IHC staining may not be available, ReceptorNet can provide accessible care. More broadly, our work shows that AI can augment physician skill sets for clinical decision-making by enabling them to leverage features too subtle for human eyes.
Additional Information
“Deep learning-enabled breast cancer hormonal receptor status determination from base-level H&E stains”, Nikhil Naik, Ali Madani*, Andre Esteva*, Nitish Shirish Keskar, Michael F. Press, Daniel Ruderman, David B. Agus, and Richard Socher, Nature Communications (2020) https://www.nature.com/articles/s41467-020-19334-3
Acknowledgments
This work is a joint effort with contributions from Nikhil Naik, Ali Madani, Andre Esteva, Nitish Shirish Keskar, Michael F. Press, Daniel Ruderman, David B. Agus, and Richard Socher.
Tissues and samples were received from the Australian Breast Cancer Tissue Bank which is generously supported by the National Health and Medical Research Council of Australia, The Cancer Institute NSW, and the National Breast Cancer Foundation. The tissues and samples are made available to researchers on a non-exclusive basis. We thank the ABCTB for their assistance in working with this data. We thank Oracle Corporation for providing Oracle Cloud Infrastructure computing resources used in this study. This research was supported in part by a grant from the Breast Cancer Research Foundation (BCRF-18-002). We thank Shubhang Desai for research assistance, Melvin Gruesbeck for help with illustrations, and Vanita Nemali and Audrey Cook for operational support. The banner image was created by Zeke Suwaratana.
References
- Naik, N., Madani, A., Esteva, A. et al. Deep learning-enabled breast cancer hormonal receptor status determination from base-level H&E stains. Nat Commun 11, 5727 (2020)
- Hammond, M. E. H., Hayes, D. F., Wolff, A. C., Mangu, P. B. & Temin, S. American Society of Clinical Oncology/College of American Pathologists Guideline Recommendations for Immunohistochemical Testing of Estrogen and Progesterone Receptors in Breast Cancer. Journal of Oncology Practice vol. 6 195–197 (2010).
- Dietterich, T. G., Lathrop, R. H. & Lozano-Pérez, T. Solving the multiple instance problem with axis-parallel rectangles. Artif. Intell. 89, 31–71 (1997).
- Ilse, M., Tomczak, J. M. & Welling, M. Attention-based Deep Multiple Instance Learning. International Conference on Machine Learning, 3376-3391 (2018).
- Putti, T. C. et al. Estrogen receptor-negative breast carcinomas: a review of morphology and immunophenotypical analysis. Mod. Pathol. 18, 26–35 (2005).
- Masood, S. Breast cancer subtypes: morphologic and biologic characterization. Womens. Health 12, 103–119 (2016).
- Dunnwald, L. K., Rossing, M. A. & Li, C. I. Hormone receptor status, tumor characteristics, and prognosis: a prospective cohort of breast cancer patients. Breast Cancer Res. 9, R6 (2007).
- Cheng, P. et al. Treatment and survival outcomes of lobular carcinoma in situ of the breast: a SEER population based study. Oncotarget 8, 103047–103054 (2017).
- Zafrani, B. et al. Mammographically-detected ductal in situ carcinoma of the breast analyzed with a new classification. A study of 127 cases: correlation with estrogen and progesterone receptors, p53 and c-erbB-2 proteins, and proliferative activity. in Seminars in diagnostic pathology vol. 11 208–214 (1994).