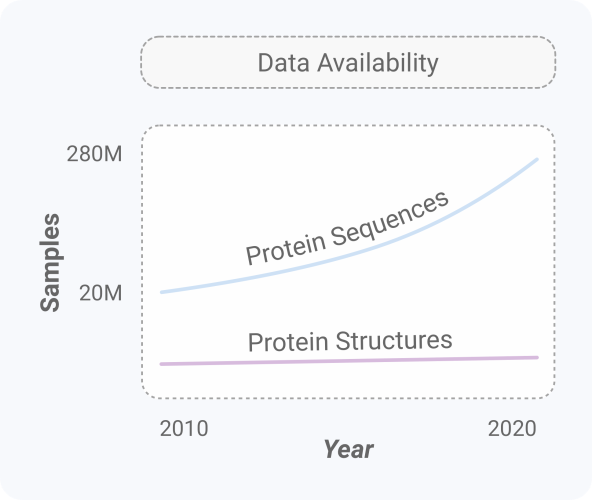
In our study [1], we demonstrate that an artificial intelligence (AI) model can learn the language of biology in order to generate proteins in a controllable fashion. Our AI system, ProGen, is a high capacity language model trained on the largest protein database available (~280 million samples). ProGen tackles one of the most challenging problems in science and indicates that large-scale generative modeling may unlock the potential for protein engineering to transform synthetic biology, material science, and human health.
Why proteins?
Let’s start with an example on everyone’s mind today. The coronavirus outbreak (COVID-19), with its contagious spread across continents and high mortality rate, has turned into a global pandemic [2] according to the WHO. To (1) better understand the coronavirus’ pathogenic nature and (2) effectively design vaccines and therapeutics, researchers across the globe are studying proteins. Within weeks, researchers were able to characterize the COVID-19 spike protein [3] which enables COVID-19 to gain entry into our human cells. Regarding detection and treatment, antibodies (also proteins) act to neutralize a virus and thereby inactivate it before causing disease.
Broadly stated, proteins are responsible for almost all biological processes critical to life. Hemoglobin carries oxygen to your cells; insulin regulates your blood glucose levels; and rhodopsin helps you see. It even extends past life itself. Proteins have been used in industrial settings to break down plastic waste and create laundry detergents.
But what is a protein?
A protein is a chain of molecules, named amino acids, bonded together. There are around 20 standard amino acids, the basic building blocks of the primary sequence representation of a protein. These amino acids interact with one another and locally form shapes (e.g. alpha helices or beta sheets) which constitute the secondary structure. Those shapes then continue to fold into a full three dimensional structure, or tertiary structure. From there, proteins interact with other proteins or molecules and carry out a wide variety of functions.
So what’s the ultimate goal of this work?
Proteins can be viewed as a language, just like English, where we have words in a dictionary (amino acids) that are strung together to form a sentence (protein). It’s impossible for us as humans to gain fluency in the language of proteins (although we dare you to try). But what if we could teach a computer, more precisely an AI model, to learn the language of proteins so it can write (i.e. generate) proteins for us? Our aim is controllable generation of proteins with AI, where we specify desired properties of a protein, like molecular function or cellular component, and the AI model accurately creates/generates a viable protein sequence.

Normally we would have to just wait for evolution, through random mutation and natural selection, to leave us with useful proteins. The emerging field of protein engineering attempts to engineer useful proteins through techniques such as directed evolution and de novo protein design [4,5]. Our dream is to enable protein engineering to reach new heights through the use of AI. If we had a tool that spoke the protein language for us and could controllably generate new functional proteins, it would have a transformative impact on advancing science, curing disease, and cleaning our planet.
In our study, we focus on modeling the primary sequences of proteins. The reason boils down to two things: (1) data scale and (2) language modeling. Advances in technology have enabled an exponential growth of protein sequences available (~280,000,000) compared to protein structures (~160,000) [6]. As machine learning is inherently data-driven, sequence modeling is a great place to start. In addition, if we view protein sequences as a language, we can leverage advances in AI and natural language processing (NLP).

What type of AI are we talking about here?
A field of artificial intelligence focusing on generative modeling has shown incredible results in image, music, and text generation. As an example, let’s take an image generation task where the objective is to create realistic image portraits of human faces. The idea is to train a high-capacity AI model (a deep neural network) on extremely large amounts of data. After sufficient training, an AI model is able to generate new facial portraits that are incredibly realistic; ones that are indistinguishable from real ones. We show a couple examples of generated images by such a model below [7].

Generative modeling has also shown remarkable success in text generation by utilizing a technique called autoregressive language modeling. At Salesforce Research, we developed CTRL [8], a state-of-the-art method for language modeling that demonstrated impressive text generation results with the ability to control style, content, and task-specific behavior. Again, it involves utilizing a high-capacity AI model trained on a large dataset of natural language. We show a couple novel pieces of text generated by CTRL below.

It’s important to underscore for both these examples in image and text generation, the model is not simply performing a search to find a relevant sample in a database. The displayed image and text above are actually generated by the AI model and do not exist in the training data.
Now for proteins, we take a similar approach to NLP by language modeling on protein sequences. Our AI model, ProGen, is given all 280 million protein sequences with their associated metadata, formulated as conditioning tags, to learn the distribution of natural proteins selected through evolution. The end-goal is to use ProGen to controllably generate a new, unique protein sequence that is functional.
But how does ProGen learn to do this?
ProGen takes each training sample and formulates a guessing game per word, more precisely a self-supervision task of next-token prediction. Let’s use an example in natural language. Imagine you were tasked with predicting the probability of the next word in the following sentence that is known to be written in a particular style in brackets:
You would expect a word such as “bats”, “lightning”, or “darkness” to have a higher probability to complete such a sentence than words such as “hello”, “yes”, or “CRMs”. Whereas if you were given the same task for the following sentence:
You would expect that words such as “love”, “sunshine”, or “happiness” to now have a higher probability. In both scenarios, we use our understanding of the previous words in the sentence (context), desired style, and English language as a whole to assign probabilities to next words/tokens.
ProGen uses this next-token prediction objective in training by formulating this game for every amino acid of all protein sequences in the training dataset for multiple rounds of training. Instead of style tags (such as horror and romance above), ProGen utilizes over 100,000 conditioning tags assigned to the protein which span concepts such as organism taxonomic information, molecular function, cellular component, biological process, and more. By the end of training, ProGen has become an expert at predicting the next amino acid by playing this game approximately 1 trillion times. ProGen can then be used in practice for protein generation by iteratively predicting the next most-likely amino acid and generating new proteins it has never seen before.
So how well does ProGen perform?
We demonstrate that ProGen is a powerful language model according to NLP metrics such as sample perplexity along with bioinformatics and biophysics metrics such as primary sequence similarity, secondary structure accuracy, and conformational energy analysis. We refer the reader to the paper for full details on the metrics description and evaluation on the held-out test set. In this post, we’ll touch on two case example proteins, VEGFR2 and GB1.
Generating VEGFR2 proteins
The protein VEGFR2 is responsible for several fundamental processes of our cells ranging from cell proliferation, survival, migration, and differentiation. We hold-out VEGFR2 protein sequences from our training dataset so ProGen never gets a chance to see them. At test time, we provide ProGen with the beginning portion of VEGFR2 along with relevant conditioning tags as input and ask ProGen to generate the remaining portion of the protein sequence.
But how do we evaluate the generation quality by ProGen? In the generative modeling examples above, we showed you image and text generations by an AI model that were visibly realistic. We need to construct an evaluation framework for a successful generation within the protein domain as well.
Again, our goal with ProGen is to generate functional proteins. VEGFR2 has a known function and known structure–in fact the full three-dimensional structure of the relevant VEGFR2 domain (at 0.15 nanometer resolution) is available. We know that structure infers function, meaning the shape of the protein gives you a strong signal as to the role of the protein. So if we can show that the ProGen generated portion maintains the structure of the protein, it strongly implies that ProGen has generated a functional protein–a successful generation!
In biophysics, there are known techniques, such as protein threading and energy minimization, that place a given amino acid sequence inside a known structure, or 3D configuration, and examine the overall energy of the protein. Like humans, proteins want to be in a relaxed low-energy state. A high energy state corresponds to the protein wanting to essentially explode indicating that you have fit the sequence to the wrong structure.
To evaluate how high of an energy is too high, we provide baselines for different levels of random mutation. For a given ground-truth (native, natural) sequence, a proportion (25-100%) of amino acids in the sequence is randomly substituted with one of the twenty standard amino acids. A 100% mutation baseline statistically indicates a failed generation. In the ideal case, we would want the energy of our ProGen sequence in the known structure to be closer to the 25% mutation or 0% mutation (native) energy levels. And that’s precisely what we show below:

Across differing generation lengths, ProGen generation quality remains steadily near native low-energy levels, indicating a successful generation. Again, ProGen is not simply performing a search within its training database. The generated sequence does not exist within the training data.
We also visualize individual samples from our experiment to examine the energy per amino acid. The ProGen sample exhibits lower energy overall, and energy is highest for amino acids that do not have secondary structure. This suggests that ProGen learned to prioritize the most structurally important segments of the protein. In the figure below, blue is low energy (stable) and red is high energy (unstable).

Identifying functional GB1 proteins
With VEGFR2, we have demonstrated the ability for ProGen to generate structure-preserving (and thereby functional) proteins from a biophysics perspective. For the protein G domain B1, named GB1, we demonstrate ProGen’s abilities with experimentally-verified functional labeled data.
Protein G is important for the purification, immobilization, and detection of immunoglobulins (antibodies)–proteins used by our immune system to neutralize pathogenic viruses and bacteria. Ideally, we would want the ability to generate GB1 proteins that are functional in terms of high binding affinity and stability. We examine a dataset [9] of 150,000 variants of GB1 by mutating four amino acid positions known to be important to overall fitness. For each one of these protein variants, the dataset reports experimentally verified fitness values which correspond to the properties that make a functional protein. Protein sequences with high fitness values are desired.
Without ever seeing the experimental data provided in the study, ProGen can identify which proteins are functional proteins. In the figure below, ProGen selected proteins exhibit a spread of high fitness values. We baseline this with the existing technique of random selection which demonstrates consistently near-zero fitness levels. This indicates that GB1 is highly sensitive to simple mutational changes and demonstrates that selecting functional GB1 proteins is a difficult task–let alone without ever training on the labeled data itself.

The intuition behind this is that ProGen has learned to become fluent in the language of functional proteins, as it has been trained on proteins selected through evolution. If given an unknown sequence, ProGen can recognize whether the sequence is coherent in terms of being a functional protein. Similar to how if you were given a string of text, you could identify if it is coherent or not based on your understanding of the English language.
What’s next?
This marks an incredible moment where we demonstrate the potential for large-scale generative modeling with AI to revolutionize protein engineering. We aim to engineer novel proteins, whether undiscovered or nonexistent in nature, by tailoring specific properties which could aid in curing disease and cleaning our planet. We hope this spurs more research into the generative space alongside existing work in protein representation learning [10-12]. Lastly, we’d love to partner with biologists to bring ProGen to the real-world. If you’re interested, please check out our paper and feel free to contact us at amadani (at) salesforce.com!
Acknowledgements
This work is done in collaboration with Bryan McCann, Nikhil Naik, Nitish Shirish Keskar, Namrata Anand, Raphael R. Eguchi, Possu Huang, and Richard Socher.
References
- Madani, Ali, et al. “ProGen: Language Modeling for Protein Generation” bioRxiv (2020).
- Chappell, Bill. “Coronavirus: COVID-19 Is Now Officially A Pandemic, WHO Says.”https://www.npr.org/sections/goatsandsoda/2020/03/11/814474930/coronavirus-covid-19-is-now-officially-a-pandemic-who-says (2020).
- Wrapp, Daniel, et al. “Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation.” Science (2020).
- Arnold, Frances H. “Design by directed evolution.” Accounts of chemical research 31.3 (1998): 125-131.
- Huang, Po-Ssu, Scott E. Boyken, and David Baker. “The coming of age of de novo protein design.” Nature 537.7620 (2016): 320-327.
- Alquraishi, Mohammed. “The Future of Protein Science will not be Supervised”. https://moalquraishi.wordpress.com/2019/04/01/the-future-of-protein-science-will-not-be-supervised/ (2019).
- Karras, Tero, Samuli Laine, and Timo Aila. “A style-based generator architecture for generative adversarial networks.” Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2019.
- Keskar, Nitish Shirish, et al. “Ctrl: A conditional transformer language model for controllable generation.” arXiv preprint arXiv:1909.05858 (2019).
- Wu, Nicholas C., et al. “Adaptation in protein fitness landscapes is facilitated by indirect paths.” Elife 5 (2016): e16965.
- Alley, Ethan C., et al. “Unified rational protein engineering with sequence-only deep representation learning.” bioRxiv (2019): 589333.
- Rives, Alexander, et al. “Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences.” bioRxiv (2019): 622803.
- Rao, Roshan, et al. “Evaluating protein transfer learning with TAPE.” Advances in Neural Information Processing Systems. 2019.